The Patient in Focus: How to Use Cell Assays for the Discovery of New Cancer Therapies
ibidi Blog | February 2, 2022 | Raúl Peña PhD, IMIM, Barcelona, Spain
Cancer is Not Cancer. Cancer Stands for Cancers, in Plural.
Under the umbrella of cancer, there are more than two hundred diseases with one unique characteristic in common: their ability to grow without stopping.
Have you ever thought about why mice are small and elephants are big? While different animals have similar cell types, mostly the same size, an elephant has millions more cells than a mouse. For example, this difference is achieved because the liver cells of mice stop dividing when the whole organ reaches a predefined size. In the case of the elephant's liver, the signal to stop reaches the cells later when compared to the mouse liver. The important fact they both have in common is that they will stop at a predetermined size, which correlates to the size of that species.
What would happen if the cells lost their ability to stop dividing? Would a mouse liver grow to be as big as the liver of an elephant? Not really, but imagine!
What is Cancer?
When your cells lose the ability to control growth (which can be influenced by many factors), it is called cancer. However, to be more accurate, the scientific and medical community uses six different, general characteristics to define cancer. We refer to them as the Hallmarks of Cancer. They specify whether or not a (random) disease should be classified as cancer. These hallmarks focus on malignant cells, and what happens to healthy cells when they get hijacked by the disease, also known as microenvironment cells.
The Aim of Cancer Research
The molecular mechanism that controls growth is called cell contact inhibition. Cell biologists define this process as the arresting of cell growth when cells come in contact with each other. In fact, most of the classic, basic cancer research focuses on how cells can skip this contact inhibition and start growing faster.
The aim behind this research is very simple to understand: If you identify the molecular mechanism that differentiates cancer malignant cells from healthy tissue cells, you will be (theoretically) able to inhibit it and, hopefully, kill the malignant cells.
Breakthroughs and Challenges in Cancer Research
Actual chemotherapeutics, and many of the antibody-based cancer therapies, are based on this strategy. Carcinogenic agents (e.g., tobacco smoke or asbestos) and even some normal cellular processes (e.g., reactive oxygen species generation as a byproduct of cellular energy production) can produce mutations in healthy cells, which affect the genes involved in cell growth. Researchers have been able to identify the key mechanisms that regulate cancer initiation and progression by comparing genomes and the protein pathways from cancer cells to healthy cells. In fact, most of the cancers today are curable, or can be controlled through the combination of surgery and drugs that can inhibit these pathways. In recent years, new treatment strategies have emerged, in combination with drug therapy, that focus on boosting the patient's own immune system by inhibiting the immune escape of cancer cells. Now, more and more cancer types can be targeted with effective (personalized) treatments.
Unfortunately, there still are cancer types that can't be successfully treated, and that means there are still thousands of patients who are struggling to fight the disease. Moreover, when a patient has been treated and cured by a certain regimen, but later suffers from a relapse, the chemotherapeutic agent that was used the first time often will not work again. To date, no treatment exists for metastasis, which is the leading cause of cancer-related deaths. Despite decades of successful cancer research, further research is still necessary. Basic research can help identify new mechanisms affecting cell growth regulation. Once identified, translational research is needed to efficiently apply these new discoveries to patients who don't yet have an effective cure.
Cellular Key Players in Cancer Development and Progression
The protein is called Snail1, and the mechanism is known as epithelial to mesenchymal transition (EMT).
EMT causes epithelial cells to lose their apical–basal polarity. This polarity loss modulates their cytoskeleton, which causes them to exhibit reduced cell–cell adhesive properties. In fact, depending on the extent of the transition, epithelial cells may individually or collectively acquire mesenchymal features, which in turn increases their motility and invasive abilities.
Particularly in the case of Snail1, the expression of this protein inside many epithelial cancer cells leads to a complete EMT, which then increases their ability to detach from other cells, as well as their motility, their invasive capabilities, and their chemoresistance (along with other cell characteristics).
So far, no basic research laboratory has been able to identify a specific treatment that can effectively block either Snail1 expression or the cell reprogramming caused by the EMT process in vivo . Moreover, as described later in this article, Snail1 expression in several non-epithelial cell types, such as fibroblasts and endothelial cells, creates activation loops that boost cancer progression, metastasis, and resistance to chemotherapy.
Methods for Investigating Cancer Cells and Drug Discovery
Cancer researchers employ different methods to study cancer development, progression, and treatment.
In our lab, for example, we use cell-based assays as the first step in identifying novel drug candidates for cancer treatments in patients. The cancer cell types and protocols used in these cell-based assays have evolved over the years. Basic protocols include the use of standard laboratory cancer cell lines. Complex assays use cancer cells isolated from a specific patient, or a mixture of cancer and non-cancer cells (e.g., stroma cells, fibroblasts), that are both present in the tumor. Cancer cells might be grown directly on plastic cultureware or, if one wants to use a more physiological, three-dimensional approach, in specific matrices that resemble those in the patient's tumor. In sophisticated assays, the cell medium includes growth factors that can also be found in the microenvironment of the cancer cells in vivo .
These assays are designed to be cost-effective, fast, simple, reproducible, and recently automated. Basically, the objective is to culture cells in the presence of a set of compounds to investigate whether these compounds effectively kill cancer cells. The main goal is to decrease cell survival in malignant cancer cells, lower cell motility in order to reduce metastatic abilities, or even achieve a complete reprogramming of cancer cells into a non-aggressive status.
Cell type reprogramming can be a difficult process to check, because expensive sequencing technologies are usually necessary. Cell motility is easier to measure when using automated systems and labware (e.g., the ibidi Culture-Insert ), which make the assays more reproducible and easier to quantify. Nowadays, the major goal of most cell-based assays that are designed to identify new therapeutic drugs is to destroy cancer cells. There are different protocols for measuring cell survival. Crystal violet or MTT stains are assays commonly used to detect survivor cells by fixing and staining them with a non-reversible dye after the treatment.
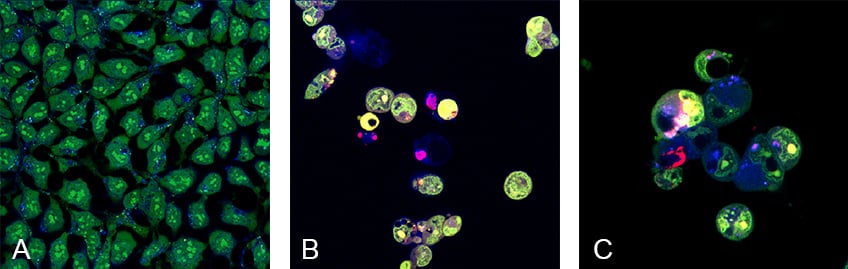
Figure 1: Visualization of cell death in Hek293 cells after chemotherapeutic treatment using in vivo staining fluorescent dyes. Cells were cultured and imaged in the µ-Plate 24 Well Black .
Acridine orange was used to stain living, healthy cells (green), and Hoechst was used to stain living cells (blue). Propidium iodide (PI) was used to stain dead or dying cells. Untreated Hek293 cells (A) do not show any dying cells in red, whereas Hek293 cells treated with the chemotherapeutic agents Doxorubicin (B), or Vinblastine (C) show a drastic increase of cell death (red) after 72 hours. Confocal microscopy with the Leica TCS SP5.
Interestingly, in some of the organelles fluorescence responds to their activity and cell viability by increasing the emission intensity or causing a dye color emission change. This allows researchers to implement automated systems to monitor drug effects. However, imaging systems require high-quality labware to ensure the optimal quality of the final image. The combination of inverted confocal microscopy and cell culture chambers or plates with a clear bottom is a state-of-the-art solution for obtaining the best imaging results, especially when using 40x or 63x objectives. The ibidi Multiwell Plates with a polymer coverslip bottom are a good alternative to glass, because they combine great cell adherence with optimal imaging properties. These imaging plates are completely flat and have an optically perfect polymer coverslip bottom that results in an excellent optical performance, even when using high magnification objectives and oil to image the cells. Moreover, black well-to-well separations block the fluorescence crosstalk during imaging. Therefore, the 96 well format ( µ-Plate 96 Well Black ) is the perfect option for testing many compounds at the same time, or the same compound in many different cell types.
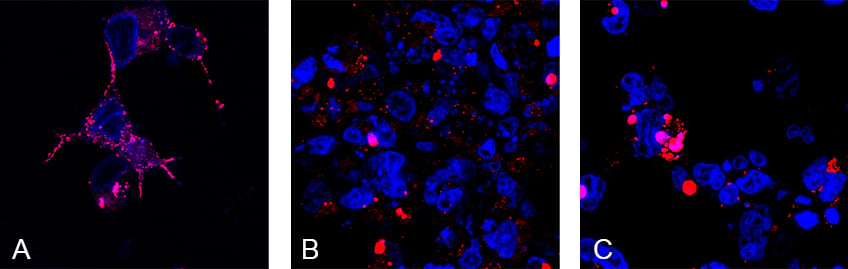
Figure 2. Live staining of mitochondria in Hek293 cells after chemotherapeutic treatment, imaged and cultured in the µ-Plate 24 Well Black
MitoTracker Red CMXRos was used to stain healthy mitochondria (red), and Hoechst was used to stain nuclei of the cells (blue). Untreated Hek293 cells (A) show high numbers of mitochondria, whereas Hek293 cells treated with the chemotherapeutic agents Paclitaxel (B), or Vinblastine (C) show a drastic decrease of mitochondria after 72 h. Confocal microscopy with the Leica TCS SP5.
However, I prefer using the 24 well format ( µ-Plate 24 Well Black ) to image my cells. They have a bigger surface for seeding and imaging higher cell numbers! It's an easy way to get cool results.
In our laboratory, cell-based assays were, are, and continue to be the initial step for many translational research projects that involve using Snail1 expressing epithelial, fibroblast, or endothelial cells. The µ-Plate 96 Well Black allows us to first perform high throughput cell viability analysis with up to 30 different candidate molecules in triplicates all at once. Next, we use the µ-Plate 24 Well Black for the detailed validation and imaging of selected candidates. When a candidate that reduces cancer cell viability, activity, or migration is identified, further in vitro research is then required to describe and characterize its underlying mechanism of action. Finally, extensive animal-based in vivo and ex vivo experiments are used to confirm the candidate's ability to block tumor growth.
But this is a story for another post…
| Raúl Peña PhD Raúl received his Bachelor's degree in Biology and his PhD in Sciences at the Basque Country University (Spain). After working in several positions at the Spanish National Research Council (CSIC), he became the senior lab manager in the lab of Antonio García de Herreros at the Institut Hospital del Mar d'Investigacions Mèdiques (IMIM) in 2007. This research group focuses on studying the role of Snail1, EMT, and the tumor microenvironment in cancer development. Raúl's own research line studies how endothelial cell activation, through the use of Snail1, promotes angiogenesis, and therefore tumor progression. Recently, the group started to focus on vascular tumors as a new model to study endothelial tumor biology. Check out Raúl's Instagram channel: @Research_Goes_Slowly |
REFERENCES
- Eduard Batlle, Elena Sancho, Clara Francí, David Domínguez, Mercè Monfar, Josep Baulida, Antonio García De Herreros. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumor cells. Nat Cell Biol. 2000 Feb; 2(2):84-9.
- David Cabrerizo-Granados, Raúl Peña, Laura Palacios, Laura Carrillo-Bosch, Josep Lloreta-Trull, Laura Comerma, Mar Iglesias, Antonio García de Herreros. Snail1 expression in endothelial cells controls growth, angiogenesis and differentiation of breast tumors. Theranostics. 2021 Jun 16; 11(16):7671-7684.
- Douglas Hanahan, Robert A Weinberg. Hallmarks of cancer: the next generation. Cell. 2011 Mar 4; 144(5):646-74.
- Jing Yang, Parker Antin, Geert Berx, Cédric Blanpain, Thomas Brabletz, Marianne Bronner, Kyra Campbell, Amparo Cano, Jordi Casanova, Gerhard Christofori, Shoukat Dedhar, Rik Derynck, Heide L Ford, Jonas Fuxe, Antonio García de Herreros, Gregory J Goodall, Anna-Katerina Hadjantonakis, Ruby Y J Huang, Chaya Kalcheim, Raghu Kalluri, Yibin Kang, Yeesim Khew-Goodall, Herbert Levine, Jinsong Liu, Gregory D Longmore, Sendurai A Mani, Joan Massagué, Roberto Mayor, David McClay, Keith E Mostov, Donald F Newgreen, M Angela Nieto, Alain Puisieux , Raymond Runyan, Pierre Savagner, Ben Stanger, Marc P Stemmler, Yoshiko Takahashi, Masatoshi Takeichi, Eric Theveneau, Jean Paul Thiery, Erik W Thompson, Robert A Weinberg, Elizabeth D Williams, Jianhua Xing, Binhua P Zhou, Guojun Sheng, EMT International Association (TEMTIA). Guidelines and definitions for research on epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2021 Dec; 22(12):834.
 (5)
(5)  (0)
(0)




