Chemotaxis Assays
A chemotaxis assay is conducted to analyze whether or not a cell type directly orients and migrates towards a defined chemoattractant. Since chemotaxis is a particularly important process during cancer development and immune infiltration, frequently used cell types in chemotaxis assays are cancer cells and immune cells.
Chemotaxis assays enable the investigation of:
- Chemotactic behavior of cells after knockout, knockdown, or overexpression of the gene of interest
- Chemotactic potential of substances
- Effect of chemotaxis inhibitors and enhancers
- Differentiation between chemotaxis and chemokinesis (enhanced migration)
- Cellular signal transduction during chemotaxis
- Changes in the cytoskeleton during chemotaxis
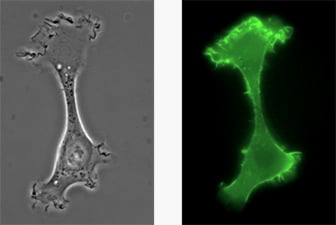
A chemotactic HT-1080 LifeAct-TagGFP2 cell migrating on a 2D surface.
2D Versus 3D Chemotaxis Assays
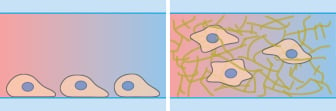
Many cells naturally grow in a three-dimensional environment. When cultured in vitro, cells are attached to a flat 2D surface and might behave differently than when they are inside a 3D gel matrix. In many cases, a 3D environment more closely resembles an in vivo situation, and this should be considered when planning chemotaxis assays.
Using the µ-Slide Chemotaxis, chemotactic gradients can easily be established in water-based gels, such as Collagen I gels and Matrigel, because the gel structure does not hinder the diffusion. Find out more information about the advantages and disadvantages of 3D chemotaxis assays here.
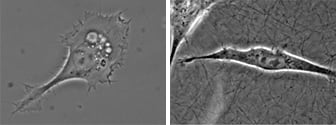
Microscopy and schematic of adherent HT-1080 cancer cells on a 2D surface (left), and embedded into a 3D Collagen I gel (right).
Chambers for Chemotaxis Assays
Several chambers with different characteristics have been developed to perform chemotaxis assays:
ibidi µ-Slide Chemotaxis
- In the ibidi µ-Slide Chemotaxis, cells migrate and can be observed in a central channel, which connects two large reservoirs
- Defined linear gradient with long-term stability
- Homogeneous cell distribution at the experimental starting point
- Ideal for live cell imaging using inverted microscopy
- Suitable for:
- 2D and 3D assays
- Fast and slow migrating cells
- Adherent and non-adherent cells
The μ-Slide Chemotaxis was developed to investigate the chemotactical behavior of fast or slow migrating, non-adherent or adherent cells on 2D surfaces or in 3D gel matrices. It is possible to observe the cell migration in linear and stable concentration profiles for more than 48 hours. As gradients are rapidly established, fast migration responses (occurring in less than 30 minutes) can also be measured.
Using the µ-Slide Chemotaxis allows for detailed and defined analysis of the migration behavior of various cell types, such as endothelial cells, fibroblasts, cancer cells, and immune cells.
Zengel P, et al. (2011) μ-Slide Chemotaxis: a new chamber for long-term chemotaxis studies. BMC Cell Biol 12:21. 10.1186/1471-2121-12-21.
read abstract
Biswenger V, et al. (2018) Characterization of EGF-guided MDA-MB-231 cell chemotaxis in vitro using a physiological and highly sensitive assay system. PLoS One 13(9):e0203040. 10.1371/journal.pone.0203040.
read abstract
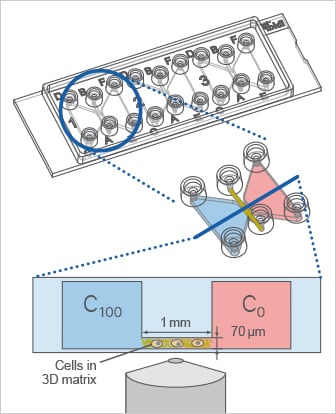
Using the µ-Slide Chemotaxis, the cells can be cultivated in a 3D gel matrix or on a 2D surface. In this example, a 3D experiment with migratory cells in a gel matrix is shown.
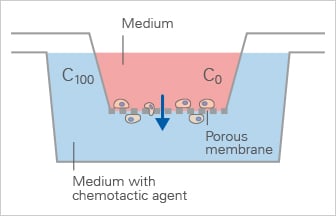
- Cells migrate from one side to the other side of a filter or a porous membrane, also called transwell cell migration assay
- Endpoint assay: when analyzing a transwell migration assay, migrating cells are counted at the back side of the membrane, and therefore cell paths cannot be analyzed
- Steep gradients
- Inhomogeneous cell distribution at the experimental starting point; cells are situated on one membrane side only
Boyden SV (1962) The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes. J Exp Med 115(3):453–66. 10.1084/jem.115.3.453.
read abstract
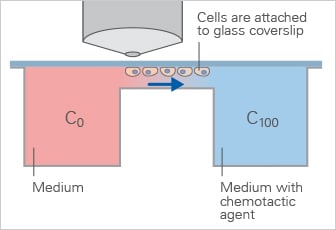
- Cells grow and migrate on a coverslip glass within a region of a bridge, which is located between two connected reservoirs
- Defined linear gradient
- No long-term stability
- Suitable for upright time-lapse microscopy
- Homogeneous cell distribution at the experimental starting point
Zigmond SH (1977) Ability of polymorphonuclear leukocytes to orient in gradients of chemotactic factors. J Cell Biol 75(2 Pt 1):606–16. 10.1083/jcb.75.2.606.
read abstract
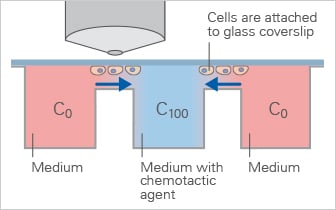
- Similar to the Zigmond chamber
- Defined linear gradient
- No long-term stability
- Suitable for upright time-lapse microscopy
- Homogeneous cell distribution at the experimental starting point
Zicha D, Dunn GA, Brown AF (1991) A new direct-viewing chemotaxis chamber. J Cell Sci 99(4).
read abstract
Read on and learn more about Chemotaxis in Cell Physiology, the Experimental Workflow of a Chemotaxis Assay, or how to do the Data Analysis of Chemotaxis Assays.




