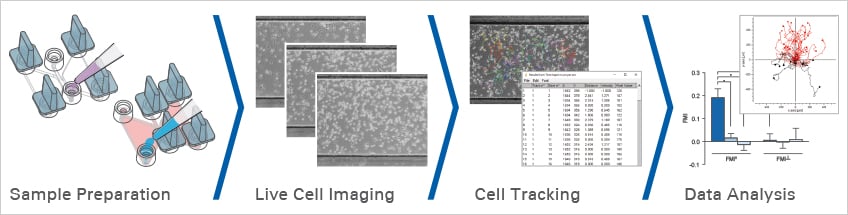
Chemotaxis
Chemotaxis refers to the directed movement of cells in response to chemical gradients, a fundamental process in various biological phenomena such as immune responses, wound healing, and cancer metastasis.
To study this behavior, chemotaxis assays are employed, allowing researchers to observe and quantify cell migration patterns under controlled conditions. These assays are crucial for analyzing cell motility, investigating disease mechanisms, and evaluating potential therapeutic agents.
ibidi facilitates these studies by offering specialized labware, such as the µ-Slide Chemotaxis, designed to create stable, reproducible gradients for high-resolution live cell imaging in both 2D and 3D environments. To learn more, explore the subchapters below, which cover key aspects of chemotaxis experiments, including gradient generation, analysis methods, and product recommendations.
ibidi Products for Chemotaxis Assays
Find more products for chemotaxis assays.
Scientific Posters
Qualitative and quantitative analysis of slowly migrating chemotactic cancer cells in 3D (PDF) Presented at the Experimental Biology (EB) Conference 2017, Chicago, Illinois, USA. | Presented at the Experimental Biology (EB) Conference 2018, San Diego, California, USA. |
Selected Publications Using Chemotaxis Assays
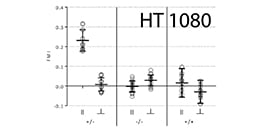
Colorectal cancer cell chemotaxis was studied using the µ-Slide Chemotaxis.
Tetrick MG, Emon MAB, Doha U, et al. Decoupling chemical and mechanical signaling in colorectal cancer cell migration. Sci Rep. 2025;15(1):4952. 10.1038/s41598-025-89152-4.
Read article
Live cell imaging of hematopoietic stem and progenitor cells was performed in the µ-Slide Chemotaxis using the ibidi Stage Top Incubator.
Prchal-Murphy M, Zehenter J, Fischer M, et al. Repurposing the prostaglandin analogue treprostinil and the calcium-sensing receptor modulator cinacalcet to revive cord blood as an alternate source of hematopoietic stem and progenitor cells for transplantation. Front Pharmacol. 2025;15:1444311. 10.3389/fphar.2024.1444311.
Read article
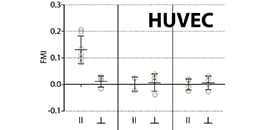
Live cell imaging of single HUVECs during chemotaxis was performed using the µ-Slide Chemotaxis.
Völkl M, Burgers LD, Zech TJ, et al. Homoharringtonine (omacetaxine mepesuccinate) limits the angiogenic capacity of endothelial cells and reorganises filamentous actin. Biomed Pharmacother. 2025;186:118025. 10.1016/j.biopha.2025.118025.
Read article
Chemotaxis of B16-F10 tumor cells was investigated using the µ-Slide Chemotaxis by generating a microenvironmental ROS gradient.
Ren X, Shi P, Su J, et al. Loss of Myo19 increases metastasis by enhancing microenvironmental ROS gradient and chemotaxis. EMBO Rep. 2024;25(3):971-990. 10.1038/s44319-023-00052-y.
Read article
The µ-Slide Chemotaxis was used to apply serum gradients across hydrogel clefts for studying 3D cell invasion.
Stöberl S, Balles M, Kellerer T, Rädler JO. Photolithographic microfabrication of hydrogel clefts for cell invasion studies. Lab Chip. 2023;23(7):1886-1895. 10.1039/d2lc01105k.
Read article
A combined 2D/3D chemotaxis assay for slow-moving cells was developed using the µ-Slide Chemotaxis.
Tomasova L, Guttenberg Z, Hoffmann B, Merkel R. Advanced 2D/3D cell migration assay for faster evaluation of chemotaxis of slow-moving cells. PLoS One. 2019;14(7):e0219708. 10.1371/journal.pone.0219708.
Read article
EGF-guided chemotaxis of MDA-MB-231 breast cancer cells was analyzed using the µ-Slide Chemotaxis.
Biswenger V, Baumann N, Jürschick J, Häckl M, Battle C, Schwarz J, Horn E, Zantl R. Characterization of EGF-guided MDA-MB-231 cell chemotaxis in vitro using a physiological and highly sensitive assay system. PLoS One. 2018;13(9):e0203040. 10.1371/journal.pone.0203040.
Read article
Application Notes for Chemotaxis Assays
Chemotaxis Assay Videos
ibidi Blog Article |
Do you want to get insight in current chemotaxis publications? Then read the ibidi Blog article 5 Methods to Analyze Chemotaxis: Recent Studies Explained.
User Protocols for Chemotaxis Assays
Frequently Asked Questions
What is chemotaxis and why is it studied?
Chemotaxis is the directed movement of cells along a chemical gradient. It is central to immune surveillance, tissue repair, angiogenesis, and metastatic dissemination. Measuring chemotaxis helps separate guidance cues from general motility changes and reveals the signaling that controls migration. Well designed assays also support screening of compounds that modulate directed cell movement.
How is chemotaxis different from chemokinesis?
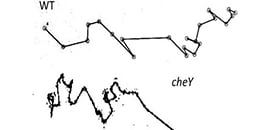
Chemotaxis describes migration with a clear directional bias toward or away from a gradient. Chemokinesis refers to changes in cell motility such as speed or persistence without a preferred direction. Distinguishing both requires controls that isolate the effect of the gradient from global activation. Using symmetric conditions alongside a defined gradient enables independent quantification of biased and random movement.
What are typical controls in a chemotaxis assay?
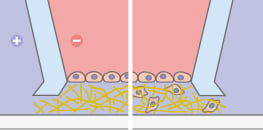
Use a negative control with no chemoattractant in either reservoir to establish baseline random motility. Use a positive control with chemoattractant in both reservoirs to reveal chemokinesis without direction. The experimental condition applies a concentration difference between the two reservoirs to create a gradient for directional testing. Comparing these three conditions supports robust interpretation and reduces confounding effects. The µ‑Slide Chemotaxis provides three independent channels, allowing all conditions to be tested simultaneously on the same slide.
When should I choose a 2D vs a 3D chemotaxis assay?
Two dimensional assays are straightforward to set up, offer high contrast, and are well suited for rapid optimization and screens. Three dimensional assays embed cells in matrices such as collagen or Matrigel to better mimic in vivo constraints including pore size, stiffness, and matrix binding. Many leukocytes and invasive tumor cells show behavior in 3D that is not captured in 2D. Choose the format that matches the biological question, available imaging hardware, and analysis pipeline.
How are stable gradients generated and how long do they last?
In diffusion based systems two reservoirs with different solute concentrations are connected by a narrow observation area where a linear profile forms. The µ-Slide Chemotaxis establishes gradients rapidly and maintains high stability for long term recordings that exceed 48 hours. Gradient linearity enables quantitative analysis and comparison across experiments. Stability also reduces user dependent variability and simplifies time lapse workflows.
What equipment is recommended for live cell chemotaxis imaging?
An inverted microscope with phase contrast or fluorescence and a stable camera is essential for time lapse acquisition. A Stage Top Incubator maintains temperature, carbon dioxide, and humidity to preserve viability and phenotype. Motorized stages and autofocus improve throughput and focus stability at multiple positions. For 3D assays choose objectives with adequate working distance and suitable immersion media.
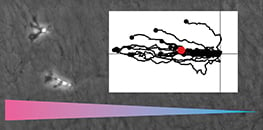
How is chemotaxis quantified?
Single cells are tracked over time to generate trajectories from which metrics are computed. Common parameters include Forward Migration Index parallel to the gradient, Forward Migration Index perpendicular to the gradient, Center of Mass, Directness, and speed. Circular statistics such as the Rayleigh test evaluate whether endpoint distributions deviate from uniformity. The Chemotaxis and Migration Tool streamlines plotting, statistics, and data export.
Can chemotaxis assays be combined with immunofluorescence or other readouts?
Yes, after live imaging samples can be fixed for endpoint immunofluorescence to assess cytoskeletal organization, receptor localization, or signaling markers. Live reporters and biosensors add dynamic information on calcium flux, actin remodeling, or kinase activity. Verify compatibility of dyes, fixatives, and mounting media with gels or surfaces. Combining functional migration data with molecular readouts strengthens mechanistic conclusions.
Which ibidi products support chemotaxis workflows?
The µ-Slide Chemotaxis provides three parallel chambers with defined and long term stable gradients suitable for 2D and 3D assays. The sticky-Slide Chemotaxis and the µ-Slide III 3in1 enable alternative layouts and gradient strategies. Stage Top Incubators maintain physiological conditions during live imaging. Collagen Type I and analysis software complete the workflow from sample preparation to quantitative data analysis.
List of pages in %s: