Example Data of a Chemotaxis Assay
We here provide all files that were created during a chemotaxis experiment for download, to give you the ability to practice every step of the analysis and interpretation of chemotaxis data.
| Experimental Setup | > | Live Cell Imaging | > | Cell Tracking | > | Data Plotting | > | Quantitative and Statistical Analysis | > | Data Interpretation |
In this example 3D chemotaxis assay with the MDA-MB-231 human breast adenocarcinoma cell line using the µ-Slide Chemotaxis, the epidermal growth factor (EGF) was used as chemotactic attractant.
Besides the experimental group (EGF/-), a negative (-/-) and a positive (EGF/EGF) control group were included. The assay was performed using three biological repeats of each experimental group.
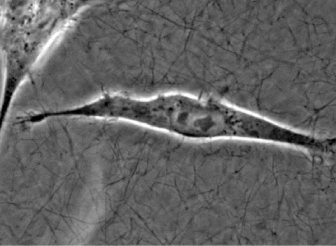
Microscopy of adherent HT-1080 cancer cells embedded into a 3D Collagen I gel in the µ-Slide Chemotaxis.
Experimental Setup
| Cells | MDA-MB-231 (human breast adenocarcinoma cells) |
| Slide | µ-Slide Chemotaxis, ibiTreat (80326) |
| 3D gel matrix | Collagen Type I, 1.5 mg/ml |
| Experimental medium | UltraCULTURETM Serum-Free Medium (Lonza) |
| Attractant medium [EGF] | 10 ng/ml EGF in UltraCULTURETM Serum-Free Medium (Lonza) |
| Chamber 1 | Experimental group (EGF/-) |
| Chamber 2 | Negative control group (-/-) |
| Chamber 3 | Positive control group (EGF/EGF) |
| Imaging mode | Phase contrast |
| Imaging period | 24 h |
| Time interval | 10 min |
| X/Y calibration (pixel size) | 0.805 µm/px |
| Number of slices/images | 145 |
| Sample size | 40 counted cell tracks per chamber, three biological repeats (one example for download) |
After live cell imaging, the following image stacks were obtained. Each stack contains 145 pictures, which were taken within a time period of 24 hours in 10 minute intervals. These images serve as raw data for subsequent cell tracking.
Download the image stacks of the example experiment here:
| EGF/- (experimental group) | -/- (negative control group) | EGF/EGF (positive control group) |
| eg_life_cell_imaging_ picture_stacks.zip | nc_life_cell_imaging_ picture_stacks.zip | pc_life_cell_imaging_ picture_stacks.zip |
The following values were obtained by manual tracking using the ImageJ Manual Tracking Plugin (tab-delimited text). The files can be directly imported into the Chemotaxis and Migration Tool for further analysis.
Download the manual tracking results of the example experiment here:
| EGF/- (experimental group) | -/- (negative control group) | EGF/EGF (positive control group) |
| eg_manual_tracking_ results.txt | nc_manual_tracking_ results.txt | pc_manual_tracking_ results.txt |
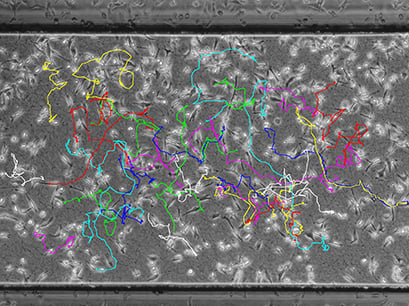
Data Plotting
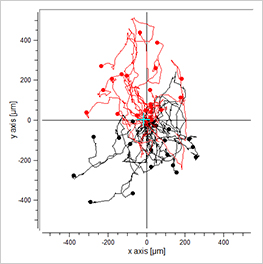
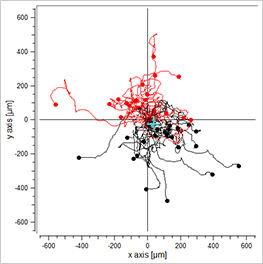
Data plotting using the Chemotaxis and Migration Tool allows for a first visual inspection of the cell trajectories, including the cell velocity and directionality:
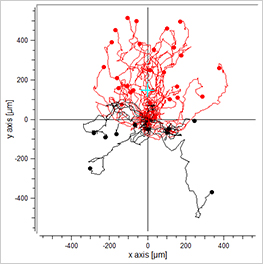
EGF/-
-/-
EGF/EGF
Visualization of the cell migration in a chemotaxis assay, showing the experimental group (EGF/-), the negative (-/-), and the positive (EGF/EGF) control group. Red cell trajectories show the migration towards the chemotactic agent, black cell trajectories indicate the migration in the opposite direction.
The EGF/- group shows cell migration in direction to the gradient. In contrast, the -/- and the EGF/EGF group show cell migration without any clear direction. In this experiment, the trajectory plots already indicate a chemotactic effect of the EGF. However, in order to confirm a chemotactic, direct cell migration towards the EGF and to exclude any environmental factors, a thorough statistical analysis of the chemotaxis parameters is mandatory.
Quantitative and Statistical Analysis
Statistical analysis of the example experiment using the Chemotaxis and Migration Tool generated the following values, which include all important chemotaxis parameters:
EGF/- (experimental group) | -/- (negative control group) | EGF/EGF (positive control group) | |
| Forward migration index, perpendicular, FMI┴ [xFMI] | 0.014 | -0.014 | 0.056 |
| Forward migration index, parallel, FMIII [yFMI] | 0.18 | -0.00021 | -0.025 |
| Directness [D] | 0.32 | 0.32 | 0.33 |
| Center of mass x [xend, µm] | 1.39 | -15.78 | 38.56 |
| Center of mass y [yend, µm] | 146.19 | 1.83 | -25.38 |
| Center of mass length [Mend µm] | 146.2 | 15.88 | 46.16 |
| Rayleigh test (p-value) | 0.0012 | 0.79 | 0.34 |
| Mean accumulated distance [daccum, µm] | 799.89 | 665.8 | 730.94 |
| Mean Euclidean distance [deuclid, µm] | 260.08 | 211.27 | 235.39 |
| Cell velocity [µm/min] | 0.56 | 0.46 | 0.51 |
As described in the Quantitative and Statistical Analysis section of the Data analysis chapter, the relevant chemotaxis parameters are used to prove the chemotactic effect of an agent.
Download the measured values of the example experiment here:
| EGF/- (experimental group) | -/- (negative control group) | EGF/EGF (positive control group) |
| eg_measured_values.txt | nc_measured_values.txt | pc_measured_values.txt |
In this experiment, the FMIII of the EGF/- group is significantly higher than the FMIII of the -/- and EGF/EGF control groups. Further, the FMIII of the EGF/- group is significantly higher than the corresponding FMI┴, which is close to zero. The FMI┴ and the FMIII of both control groups are close to zero as well. The Rayleigh test results in p < 0.05 in the EGF/- group and p > 0.05 in the control groups.
Taking all these criteria into account, the data analysis shows a significant chemotactic migration of the MDA-MB-231 cells in direction of the EGF.
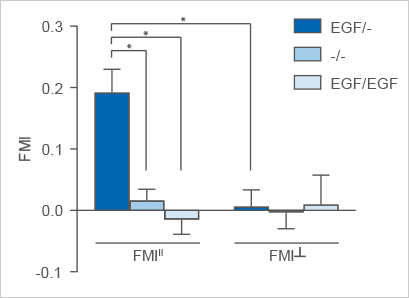
Averaged FMI II and FMI ┴ of the EGF/- experimental group in comparison to the -/- and the EGF/EGF control groups. Significant chemotaxis is observable parallel to the gradient in the EGF/+ group. Please note: this graph includes three biological replicates.
Read on and learn more about the Data Analysis of Chemotaxis Assays or the Experimental Workflow of a Chemotaxis Assay.




