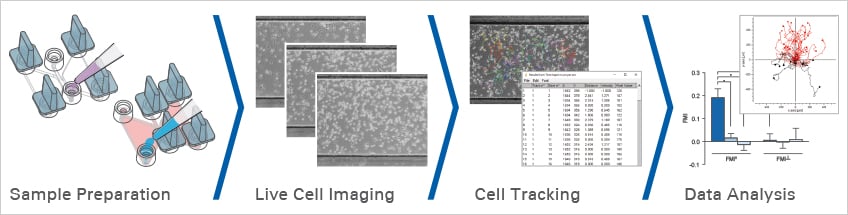
Experimental Workflow of a Chemotaxis Assay
Before Starting
Necessary Equipment
Basic requirements
- Inverted phase contrast microscope (5x or 10x objective recommended)
- Camera for time lapse image acquisition
- Stage Top Incubator (required for most mammalian cell types)
Recommended extensions
- Motorized stage for parallel image acquisition
- Autofocus
Questions to Ask
For a successful chemotaxis experiment, please address the following questions before starting.
Sample Preparation
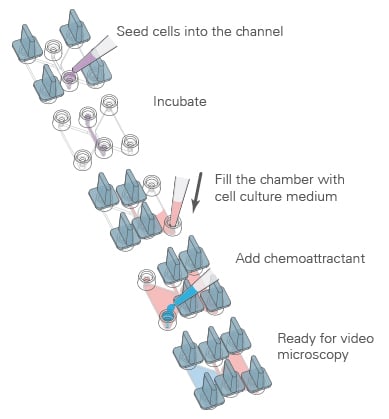

Procedure
Using the µ-Slide Chemotaxis, reproducible chemotaxis assays with defined chemotactic gradients can be carried out.
First, the cells are seeded, which can be done either in a 2D or a 3D environment. After incubation and cell attachment, the two reservoirs of each chamber are filled with the chemoattractant, according to the determined loading scheme.
More information about the detailed handling of the µ-Slide Chemotaxis can be found in the Application Note AN 17: Chemotaxis 2D and 3D (PDF).
ibidi Solutions

The µ-Slide Chemotaxis contains 3 separate chambers for parallel chemotaxis assays using slow or fast migrating cells.
It allows for the creation of a precisely defined, stable chemotactic gradient in a reproducible environment.
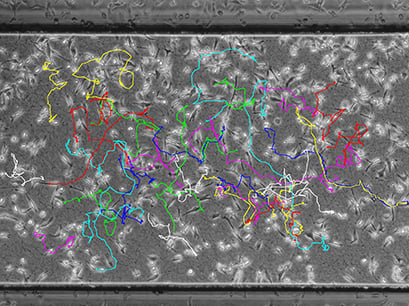
Live Cell Imaging
Time-lapse microscopy of the migration of MDA-MB-231 human breast adenocarcinoma cells in a collagen gel. Cells were observed over a period of 24 hours in the µ-Slide Chemotaxis with EGF as chemotactic agent. Physiologic conditions were maintained by the ibidi Heating and Gas Incubation System.

Procedure
Live cell imaging under physiologic conditions enables for the detailed documentation of the cell migration over time, a process which is necessary for the proper analysis of a chemotaxis assay.
The duration of the imaging period depends on the cell type (e.g., fast migrating leukocytes or slow migrating tumor cells or fibroblasts) and the environmental conditions (e.g., type and concentration of the chemotactic agent). A typical video microscopy setup for slow migrating cells includes taking a photo every 2.5–10 minutes over a period of 24 hours.
Since each experiment optimally contains tracking data from 20–40 single cells, low-magnification microscopy objective lenses, such as 4x or 10x, should be used.
ibidi Solution

The ibidi Stage Top Incubators provide a physiological environment under the microscope, which enable live cell imaging during short-term and long-term chemotaxis assays.
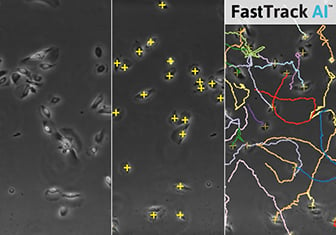
Cell Tracking
Visualization of cell traces after tracking with the ImageJ Manual Tracking plugin.
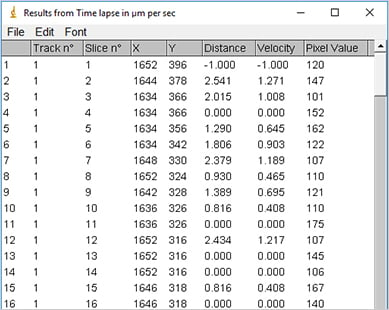
Output of the ImageJ Manual Tracking Plugin after tracking; data table with positional values of each tracked cell (x, y) for each time point (t).

Procedure
Typically, live cell microscopy of a chemotaxis assay creates a temporal image stack, where each image displays the exact cell position at a specific time point.
For quantification of their movement, single cells are tracked over time by determining their position on each frame of the image stack. The tracking can be done either manually or automatically using special software. Afterwards, the positional values of each tracked cell (x, y) are available for each time point (t) and can be further analyzed.
The ibidi Science HubDiscover our application chatpers and free online courses. |
|
Manual trackingThe ImageJ Manual Tracking Plugin assists in manually tracking cell migration in chemotaxis assays. It can be downloaded for free here. |
|
Data Analysis
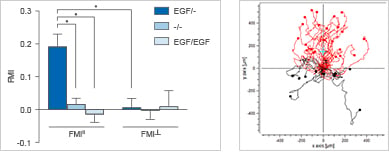
![]()
Find a detailed description of the data analysis steps of chemotaxis assays here:
Questions to Ask Before Starting an Experiment
In order to set up a chemotaxis assay correctly, it is crucial to answer the following questions first:
Which cell type are you planning to investigate?
Knowing your cell type of interest—including the culture medium requirements and the migration speed—is essential for subsequent assay planning.
What is the migration speed / velocity of the cell type being analyzed?
Whereas some cells have a low migration speed (e.g., tumor cells or fibroblasts), other cell types (e.g., leukocytes) migrate very quickly. The cell migration speed will determine the duration of the experiment and the intervals required between the images. Also, the gradient stability must be high enough to fit the total duration of the experiment. The µ-Slide Chemotaxis is suitable for chemotaxis assays with both slow and fast migrating cells.
| Slow migrating cells | Fast migrating cells | |
| Migration type | Mesenchymal | Amoeboid |
| Examples | Endothelial cells, cancer cells, fibroblasts, stem cells | T cells, neutrophils, dendritic cells, Dictyostelium discoideum |
| Speed | One cell length per hour: ∼10 µm/h | One cell length per minute: ∼10 µm/min |
| Frame rate (time lapse) | 1 image per 10 min | 1 image per 30 s |
| Typical experiment duration | 12–24 h | 15–30 min |
What is the optimal culture medium?
Most cell types are cultured in a medium supplied with fetal calf serum (FCS). FCS contains many factors (e.g., enzymes, hormones and growth factors) that can influence the cell migration, and therefore might alter the outcome of a chemotaxis assay. For example, the effects of a chemotactic agent on cell migration can be masked by FCS-induced effects. Gradual reduction of the FCS concentration in the medium, before starting the assay, is one way to overcome this issue. Furthermore, special, serum-free culture media without FCS have been developed. The optimal medium composition for each cell type of interest must be tested before starting the assay.
What is the optimal seeding density for the cell type of interest?
The seeding density depends on many cellular factors, such as the proliferation rate, behavior, shape, epithelial or mesenchymal state, and the dependence on cell-cell contacts. In addition, the experiment duration must be considered when determining the optimal cell seeding density. In order to have enough trackable cells, the density must not be too low when starting the experiment. At the endpoint of the experiment, single cells should be clearly definable and trackable. Considering these factors, the optimal seeding density should be separately determined for each cell type before starting the chemotaxis assay.
How many experiments should be performed?
Typically, three to five repeat experiments are sufficient to create significant data from the chemotaxis group and the respective control group. Each experiment should contain tracking data from 20–40 single cells, which is possible using low-magnification microscopy objective lenses, such as 5x or 10x.
Should I perform a 2D or a 3D chemotaxis assay?
Most cells are naturally embedded in a 3D matrix. Culturing them in a 2D environment during a chemotaxis assay might alter their behavior and migrational capabilities. To overcome this issue, cells can be embedded in a 3D matrix that mimics their natural environment, such as collagen, Matrigel, or other hydrogels. The µ-Slide Chemotaxis is ideally suitable for both 2D and 3D experiments.
Advantages of 3D Chemotaxis Assays
- More in vivo-like setting for most cell types
- Highly defined environment (e.g., fibers or matrix)
- Chemotaxis assays with suspension cells possible
Limitations of 3D Chemotaxis Assays
- Difficult gel handling; more parameters to control during the experiment
- Cells might attach to 2D surface, thus creating 2.5D conditions
- Cells might go out of focus during 3D tracking
Find more information about 2D and 3D chemotaxis assays in the following Application Notes:
Which controls must be included in the experiment?
For the correct analysis of a chemotaxis experiment, it is crucial to include a negative control without any chemoattractant (-/-), as well as a positive control with chemoattractant over the entire chamber (+/+).
Using the µ-Slide Chemotaxis, a minimum of control measurements are required, since all conditions other than the gradient are symmetric. This allows for the analysis of chemotaxis and chemokinesis independently of each other. In the example, the chemoattractant induces both the chemotaxis and chemokinesis of cancer cells.
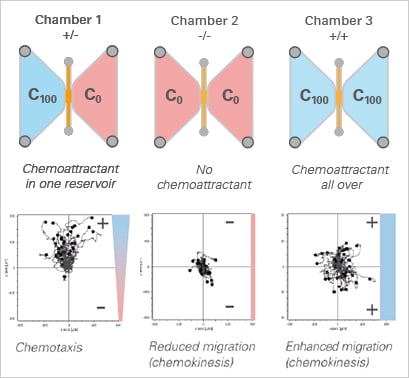
Experimental setup (top) and data plots (bottom) of a chemotaxis assay including the experimental group (+/-), a positive (+/+), and a negative (-/-) control group.
Read on and learn more about Chemotaxis in Cell Physiology, different Chemotaxis Assays, or how to do the Data Analysis of Chemotaxis Assays.