Tumor Microenvironment
and Immunooncology
Solid tumors are surrounded and interact with the tumor microenvironment (TME), which is composed of extracellular matrix, fibroblasts, immune cells, lymphatic and vascular endothelial cells, and further stroma cells. Contrasting the classical tumor cell-oriented basic research, the TME has gained more and more attention in recent decades, opening new perspectives for therapeutic approaches (Balkwill et al., 2012).
![]()
Hypoxia is common in cancer cells and the TME as a result of excessive tumor growth and insufficient vessel connection and oxygen supply. It can influence cancer biology in many ways, promotes angiogenesis, and correlates with cancer progression and a poor prognosis. Therefore, when working with cancer cells or stromal cells, researchers should consider working under hypoxic conditions, as it reflects the physiological oxygen levels in tumors more accurately. The ibidi Stage Top Incubators can mimic hypoxic conditions on every inverted standard microscope during live cell imaging experiments.
Many immune cell types, such as T cells and myeloid cells (e.g., tumor-associated macrophages, TAMs), are part of the TME and have already been intensely investigated for their therapeutic suitability. Depending on the cell subtype, they can have both pro- or anti-tumor activity and be associated with a good or bad prognosis. Especially T cells harbor the potential to be used to destroy tumor cells in a patient-specific approach: T cells with engineered chimeric antigen receptors (CAR T cells) have been developed for adoptive T cell transfer therapy and showed clinical success for the treatment of B cell lymphomas.
In a healthy system, the immune checkpoints limit the immune response to prevent collateral tissue damage. However, in a tumorigenic environment, cancer cells can control these checkpoints to inhibit their own destruction by the immune system. The clinical approval of immunotherapy of specific cancer types, such as melanoma, uses an entirely new tumor treatment approach that blocks immune checkpoints (e.g., by anti-CTLA-4, anti-PD-1, or anti PD-L1) and has improved the outcome of thousands of patients. However, not all patients benefit from the immunotherapy, with some not responding at all or others gaining resistance. This implies that additional basic research is needed to evaluate a more patient-specific treatment strategy (Topalian et al., 2015; Waldman et al., 2020).
Various in vitro approaches exist for deciphering the communication and interaction mechanisms between tumor cells and their microenvironment: CAR-T cell killing assays, co-culture of cancer cells and immune cells (or other stromal cells), and rolling and adhesion assays, just to name a few, help researchers worldwide to gain knowledge of this important field of cancer research.
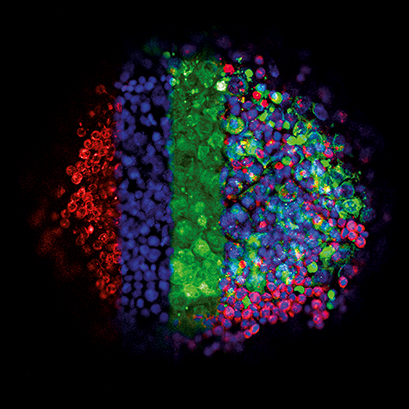
This image shows the three-dimensional growth of germ cell tumor cells (green, GFP) and immune cells (lymphocytes, red, mCherry) using the hanging drop technique. Cell nuclei appear in blue (DAPI). In 3D culture, a close interaction between the diverse cell types develops. The drops were transferred into an ibidi μ-Plate 96 Well. The image was acquired using a Zeiss LSM 710 confocal microscope with a 10x objective. Data by Gillian Ludwig, Daniel Nettersheim, Translational Urooncology, Department of Urology, University Hospital Düsseldorf, Germany.
Balkwill, F. R., Capasso, M., & Hagemann, T. (2012). The tumor microenvironment at a glance. Journal of Cell Science. 10.1242/JCS.116392
Read article
Topalian, S. L., Drake, C. G., & Pardoll, D. M. (2015). Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 10.1016/J.CCELL.2015.03.001
Read article
Waldman, A. D., Fritz, J. M., & Lenardo, M. J. (2020). A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nature Reviews Immunology. 10.1038/s41577-020-0306-5
Read article
Experimental Example: Automated Analysis of CAR-T Cell Killing Assays
CAR-T cells represent a promising new cancer therapy tool. Live cell imaging allows real-time T cell/cancer cell interaction analysis with single cell resolution. However, analysis of confluent cell layers is time-consuming and, therefore, impossible in high throughput screens. To facilitate high throughput label-free analysis of T cell potency in a live cell imaging setup, we generated arrays of homogenously distributed cancer cells using the ibidi Micropatterning technology. By combining optical analysis and advanced image processing, cytotoxic T cell activity over time on a single cell level can be evaluated without the use of any labeling.
Single Cells in a 2D Environment
Time lapse microscopy of a CAR-T cell killing assay with RCC-26 renal tumor cells and JB4 T cells on a single cell pattern (µ-Slide VI 0.4, 20 µm squares, 120 µm distance, rectangular). Data were analyzed using FastTrack AI by MetaVi Labs.
Multi-Cell Spots in a 3D Collagen Matrix
RCC-26 renal cancer cells immobilized on multi-cell pads (µ-Slide VI 0.4, 200 µm circles, 600 µm distance, hexagonal). Effector T cells applied in a collagen I matrix (Collagen Type I, Rat Tail) induce apoptotic body formation of cancer cells.
ibidi Solutions for Immunooncology and Analysis of Tumor Microenvironment
The ibidi µ-Slides and µ-Dishes include different geometries that combine optimal conditions for functional cell-based assays using cancer cells, immune cells, or other stromal cell types. They are ideal for immunofluorescence, live cell imaging, and high-resolution microscopy. The ibidi labware is available with the ibidi Polymer Coverslip and the ibidi Glass Coverslip. For high-throughput experiments, we offer the ibidi µ-Plates, which are available with 24 Wells or 96 wells |
|
The ibidi Stage Top Incubators provide physiological conditions for live cell imaging on every standard inverted microscope. They include CO2 and O2 control (e.g., for hypoxia experiments) as well as actively controlled humidity. They are available for single slides, dishes, and multiwell plates. |
|
µ-Slides With Single-Cell µ-Pattern are ready-to-use micropatterned slides with ideal spacing for single cell assays. The µ-Slides With Multi-Cell µ-Pattern enable spatially defined cell adhesion for spheroid and organoid generation. Both solutions have been optimized for long-term culture and high-resolution imaging and are useable for versatile cell-cell interaction studies (e.g., a CAR-T cell activity assay) and much more. |
|
The ibidi Pump System is ideal for long-term cell culture under flow with defined shear stress values and is compatible with all µ-Slides with Luer adapters. It simulates defined continuous and pulsatile laminar flow, and oscillatory flow to study cells in a more physiological environment. It is optimal for rolling and adhesion assays, transmigration, and invasion studies. Also, cells, spheroids, and organoids can be perfused for optimal nutrition. |
|
ibidi provides a variety of Channel Slides with different geometries. The µ-Slide I Luer family have one channel designed for standard flow experiments and rolling and adhesion assays. The µ-Slide VI has 6 channels ideal for parallel flow assays. Both are available with the ibidi Polymer Coverslip and the ibidi Glass Coverslip, plus different heights and coatings. |
|
The μ-Slide I Luer 3D allows for creating an endothelial barrier without the need for an artificial filter membrane. Endothelial cells can be seeded on a suitable gel matrix, such as Collagen Type I. After connecting the slide to a pump and applying defined shear stress, an in vivo-like endothelial barrier is created, which is useful for rolling and adhesion assays or transendothelial migration studies. |
|
The µ-Slide Chemotaxis and the sticky-Slide Chemotaxis are ideal for analyzing single cell migration of immune cells and cancer cells in 2D and 3D, because they quickly create a stable chemotactic gradient. These gradients are easily established in 2D or water-based 3D gels, such as Collagen I gels and Matrigel® because the gel structure does not hinder the formation of a soluble gradient by diffusion. |
|
Selected References
Review stating the use of the ibidi Pump System as a microfluidic-based method for mimicking and analyzing the tumor microenvironment
Crouigneau R, Li YF, Auxillos J, et al. Mimicking and analyzing the tumor microenvironment. Cell Rep Methods. 2024;4(10):100866. doi:10.1016/j.crmeth.2024.100866
Read article
3D chemotaxis assays of CD8+ peripheral blood T cells (PBTs) from patients with head and neck squamous cell carcinoma (HNSCC) using the µ-Slide Chemotaxis
Newton, H. S., Gawali, V. S., Chimote, A. A., Lehn, M. A., Palackdharry, S. M., Hinrichs, B. H., Jandarov, R., Hildeman, D., Janssen, E. M., Wise-Draper, T. M., & Conforti, L. (2020). PD1 blockade enhances K+ channel activity, Ca2+ signaling, and migratory ability in cytotoxic T lymphocytes of patients with head and neck cancer. Journal for Immunotherapy of Cancer. 10.1136/JITC-2020-000844
Read article
Co-culture assay with tumor cells and macrophages in a breast cancer model using the µ-Dish 35 mm, high
Sharma, V. P., Tang, B., Wang, Y., Duran, C. L., Karagiannis, G. S., Xue, E. A., Entenberg, D., Borriello, L., Coste, A., Eddy, R. J., Kim, G., Ye, X., Jones, J. G., Grunblatt, E., Agi, N., Roy, S., Bandyopadhyaya, G., Adler, E., Surve, C. R., … Oktay, M. H. (2021). Live tumor imaging shows macrophage induction and TMEM-mediated enrichment of cancer stem cells during metastatic dissemination. Nature Communications. 10.1038/s41467-021-27308-2
Read article
Co-culture assay with bone marrow-derived macrophages (BMDM) and tumor cells in the µ-Plate 96 Well Black
Luthria, G., Li, R., Wang, S., Prytyskach, M., Kohler, R. H., Lauffenburger, D. A., Mitchison, T. J., Weissleder, R., & Miller, M. A. (2020). In vivo microscopy reveals macrophage polarization locally promotes coherent microtubule dynamics in migrating cancer cells. Nature Communications. 10.1038/s41467-020-17147-y
Read article